As previously communicated in my last post, I am currently working on a Dekkera anomala proteome (strain YV396) to get a better understanding of the pathways associated with compounds found in beers like Lambics. I compiled my data and first results into a easier to read format (see below). In case there is something else of interest in the genome, I will likely publish these results on my blog as well.
Category Archives: Projects
Insight into the Dekkera anomalus YV396 genome – part 1
It has been a while since I look at Brettanomyces genomes since there wasn’t that much data available to play around. And mainly B. bruxellensis data is available due to its importance in the wine industry. This all changed when I came across the deposited draft genome assembly of a Dekkera anomalus YV396 genome in June by et al. Since there was no annotation material available for this genome I quickly decided to give the annotation a shot myself. Simply because I am interested in certain pathways in Brettanomyces. Everything I needed was my Ubuntu notebook (which died during the annotation process), my new Ubuntu workstation (replacing the notebook) and some Python coding. No access to a cluster whatsoever. Is it possible to finish an entire annotation project at home? You will find out very shortly. As I am still compiling data for a post, I want to start sharing the material part as well as the first abstract. Just to give you a sneak-peek into the project. The remaining part of the genome & proteome project will get published very soon. Just give me some additional time to finish up the various pathway analysis and writing up the paper. Still a lot to be discovered in the new genome…
I – Methods
Genome assembly
The draft genome assembly of Dekkera anomalus strain YV396 (isolated from a Belgian brewery) was retrieved from GenBank (accession number LCTY00000000.1; June 2015) deposited in May 2015 by KU Leuven [ et al.]. Illumina HiSeq data (100x coverage) was assembled into a genome using SOAPdenovo v.1.05. The statistics for the obtained assembly are summarized in Tab. 1.
 Gene prediction
Gene prediction
Gene prediction on contigs was performed using the AUGUSTUS web-service (AUGUSTUS parameter project identifier: pichia_stipitis, UTR prediction: false, report genes on both strands, alternative transcripts few, allowed gene structure: predict any number of (possibly partial) genes, ignore conflicts with other strand: false) [Stanke et al.2006, 2008]. The gene prediction statistics are summarized in Tab. 2.
 Gene annotation
Gene annotation
Gene annotation was performed by Blast2GO including remote blastx on NCBI and InterProScan for domain predictions [Conesa et al, 2005]. GO-term mapping and annotation performed by Blast2GO pipeline. Close to 3,000 out of the predicted 4,160 could be annotated by Blast2GO (Fig 3). Another subset of about 600 sequences could be mapped to a biological function without a GO term and about 460 sequences only resulted in BLAST hits which could not be further associated with a protein function.
 Most abundant species associated with the best blastx hits were Dekkera bruxellensis, Ogataea polymorpha and Pichia kudriavzevi (not shown).
Most abundant species associated with the best blastx hits were Dekkera bruxellensis, Ogataea polymorpha and Pichia kudriavzevi (not shown).
References
- Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO:a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21(18):3674–3676.
- Stanke, M., Diekhans, M., Baertsch, R., and Haussler, D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics, 24(5):637–644.
- Stanke, M., Schoffmann, O., Morgenstern, B., and Waack, S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics, 7(1):62.
- (2015) Purification and characterization of a novel Brettanomyces anomalus beta-glucosidase enzyme suitable for food bioflavoring – unpublished.
Sugar composition of wort
Short one for today. I would like to share some information about the sugar composition of wort since I had to take this into consideration for an upcoming project I am preparing for publishing here soon (yes, its yeast related). Lets talk sugars today!
I am not sure how many homebrewers thought about the actual sugar composition of their wort before. And I am not speaking about fermentable and non-fermentable ones. The real composition like sucrose, maltose, glucose etc. The question now is why one might think about that problem in the first place. For example, if you are interested to know if a non-Saccharoymces yeast (capable of fermenting glucose only) can ferment something in a wort, you might need to know if glucose is even present in the first place (and this example is pretty close to the question I asked myself to eventually investigate the composition of sugars in wort).
The composition of sugars in wort has been addressed a couple of years ago and published in various papers. Like “Determination of the sugar composition of wort and beer by gas liquid chromatography” by Otter et al published in 1967 [get me to the paper]. I will not go into the scientific details as well as experimental setup of this paper but would like to discuss the results.

Fig 1: Paper header
Otter et al determined the concentrations of six sugars (fructose, glucose, sucrose, maltose, maltotriose and maltotetraose) in 15 different worts with various OGs (ranging from 1.027 up to 1.093). I averaged the sugar compositions of the 15 samples as amount of sugar X relative to the total amount of sugar present in wort. Just to give me a rough idea. So don’t read too much into the numbers here. It’s about the ratio or more like sugar X is highly abundant or not. And yes, I thought about effects of grain bill composition, mash schedules, mash pH, you name it on the sugar composition. Getting a rough idea here.

Fig 2:Sugar composition w[%] of total sugar as average of 15 different worts
About half of the sugars present in wort is maltose (Fig 2). Followed by maltotriose and glucose. And some smaller amounts of fructose, sucrose and maltotetraose. Maltotetraose by the way is a dextrin and can be counted as non-fermentable. Standard Saccharomyces cerevisiae strains are capable of fermenting all the present sugars except maltotetraose. Which might explain why S. cerevisiae is the working horse of brewers. In summary, maltose makes up about half of the total sugars followed by glucose and maltotriose. And some minor amounts of fructose, sucrose and maltotetraose. I am actually surprised about the amount of glucose present in wort. I did not expect that at all.
So there you go. I will address the initial problem about a specific sugar metabolism of a non-Saccharomyces yeast in a future post including some empirical data. Stay tuned!
Brettanomyces Phylogenetic II & WLP Brett Trois crisis
Starting 2015 with science. This is a more in depth post about yeast phylogeny. Please have a look at the more basic post here. This time, I would like to discuss the relationship of various Brettanomyces/Dekkera strains and share my results concerning the recent WLP Brettanomyces bruxellensis Trois yeast ID crisis.
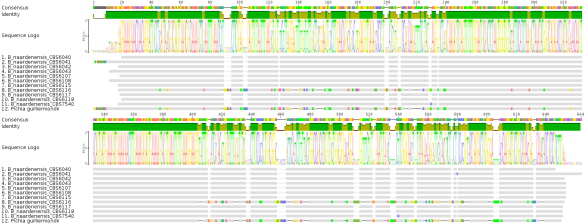
I would like to start with the Brettanomyces/Dekkera strains first. I obtained 38 Dekkera/Brettanomyces sequences (26S rDNA) from the CBS database, aligned them using MUSCLE (run in default mode, see MSA in Fig 1) and reconstructed a phylogenetic tree using MABL (model HKY85) and rendered using TreeDyn (run in default mode, Fig 2).
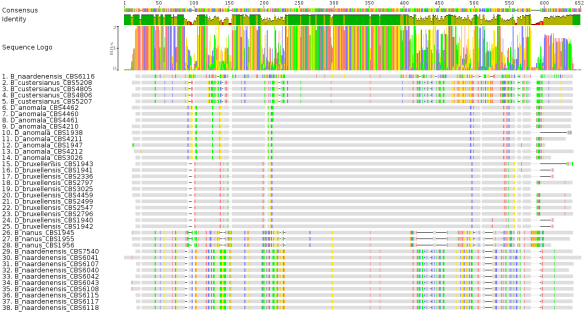
Fig 1: MUSCLE multiple sequence alignment (MSA) of 26S rDNA sequences from 38 different Brettanomyces/Dekkera strains obtained from CBS
I would like to emphasize the importance of having a look at the intermediate steps in constructing a phylogenetic tree since sequence alignments can happen in various ways. Looking at the MUSCLE output, one can already expect to see the individual strains clustered together due to their sequence similarities. However, there is one particular sequence that seems to be a bit off (sequence number 1, B. naardenensis CBS 6116). This sequence seems to be different from all the other B. naardenensis sequences shown at the bottom (sequences 29-38). Based on this result, one can expect to see CBS 6116 to be an out-group to the other B. naardenensis sequences. In summary, the alignments seem to be okay and let’s have a look at the phylogenetic tree of the 38 sequences.

Fig 2: Brettanomyces/Dekkera phylogenetic tree using 26S rDNA sequences obtained from CBS. Branch length is proportional to the number of substitutions per site (values shown in red)
As already observed in the MSA, the individual Brettanomyces/Dekkera species cluster together as expected (Fig 2). B. naardenensis CBS6116 does indeed form an out-group (isolated from lemon drink in France) and is kind of distant to the other B. naardenesis sequences. So why is this CBS 6116 sequence different?
To address this question, I tried to figure out first what other sequences are similar to CBS 6116 by BLASTing against the NCBI nr database (run in default mode). I got several hits and manually inspected the results via alignment. The CBS 6116 sequence has high sequence identities to Pichia guillermondii (Fig 3). One might reason – based on these results – that CBS 6116 is not a Dekkera/Brettanomyces strain.
Going back to the phylogenetic tree in Fig 2. It seems that all the current Dekkera/Brettanomyces species end up in the same clusters and show enough substitutions to tell individual species apart. This is very important information for everyone interested in determining the species of an unknown Dekkera/Brettanomyces strain.
WLP Brettanomyces Trois ID crisis
I got interested in the WLP story (WLP Brettanomyces bruxellensis Trois not being a Brettanoymces strain) and started by looking at the WLP Brett Trois ITS sequences I received from Omega Labs (reads 64-ITS1.ab1 and 64-ITS4.ab1). I had a quick look at the chromatograms and would not use any of the reads for my own work due to its low base quality and multiple peak calls at certain positions. Besides the read from Omega Labs, I received a read used to ID the B. Trois as Saccharoymces cerevisiae at Charles River’s Lab as well as the read published by SuiGeneris. Since I don’t have the chromatograms for these two sequences, I manually inspected the reads in various alignments to get an idea about their quality. Lets start with my analysis for the WLP Brettanoymces bruxellensis Trois ID’ing based on phylogenetic trees.
To get the most likely phylogenetic tree one has to follow some basic rules. Beginning with looking at the same shared derived homologous traits (homologous DNA sequences) and verifying that no other DNA sequence alterations impact the phylogenetic tree like sequencing errors (wrong base calls, no base call, multiple base calls etc). So far so good. I aligned some ITS2 regions from various CBS Saccharomyces strains to the various ITS2 reads from WLP’s Brettanomyces Trois (Fig 4).

Fig 4: MSA of ITS2 rDNA sequences from various Saccharomyces sp. strains obtained from CBS and ITS2 sequences from WLP B. Trois
Lets have a look at the alignment shown in Fig 4. Especially at the 64-ITS4.ab1 read from WLP B. Trois (sequence 1 shown at the top). One can easily see, that various base differences exists compared to the other sequences in the alignment. Supporting the initial idea that the DNA sequence from this read is not very reliable nor very representative (if one compares the read to the two other WLP B. Trois reads shown at the bottom). Due to the differences in this read, I would not be surprised to see this read out-grouped in the phylogeny tree later on. The two reads from Charles River’s Lab and SuiGeneris seem to be way better and similar to other Saccharomyces sp. sequences shown in the alignment.
To make the phylogeny more computational efficient and more reliable, I extracted the sequences mapping to the two WLP sequences (sequence regions from the right side in Fig 4) to receive the alignment shown in the next figure (Fig 5).

Fig 5: MSA of ITS2 rDNA sequences from various Saccharomyces sp. strains obtained from CBS (extracted)
A first look at the alignment in Fig 5 reveals some hotspots for variations like gaps and different base calls (color regions). The question I would like to address now is whether the variations are due to speciation or artifacts. Artifacts are commonly more random than variations due to speciation. A quick inspection reveals lots of random variations in the 64-ITS4 read but none/few for the two other WLP B. Trois reads.
The phylogeny tree obtained for the sequences shown in the alignment in Fig 5 is shown in Fig 6

Fig 6: Saccharomyces phylogenetic tree using ITS2 sequences obtained from CBS & WLP B. Trois from CharlesRiverLab/SuiGeneris and n64-ITS4.ab1 from Omega Lab. Branch length is proportional to the number of substitutions per site (values shown in red)
As expected the n64-ITS4 read gets out-grouped and might be interpreted as a different species. Well this is true if one just looks at the tree in Fig 6 but did not look at the previous alignments and possible reasons for the out-grouping. In this case, the quality of the base calls for the n64-ITS4.ab1 read are very poor and led to wrong base calls after all (investigated by pair-pair alignments). In summary, the differences leading to an out-grouping of WLP B. Trois based on the n64-ITS2 read are not due to the physical differences in the DNA sequence but due to sequencing errors.
In summary, the two WLP B. Trois sequences group together with other Saccharomyces cerevisiae strains. Supporting the current view that WLP B. Trois might indeed not be a Dekkera/Brettanomyces strain (at least based on ITS2 sequence homology). Or at least the samples of WLP B. Trois that float around these days.
As I do isolate and work with my own Brettanomyces strains, it happened to me various times that I was able to observe some Saccharomyces cerevisiae beside the initial Brettanomyces strain after some serial re-pitches (and I did not use Saccharomyces for the fermentation). This Saccharomyces contamination might lead to problems during the propagation. Saccharomyces cerevisiae might overgrow Brettanomyces increasing the Saccharomyces:Brettanomyces ratio even further. Eventually leading to very few Brettanomyces cells left in a population. Keeping Brettanomyces samples is not as easy as one might think. And I would not be surprised if more yeast conundrums turn up in the next years.
Launching Eureka Brewing’s Yeast lab
Eureka, I am really pleased to announce that I finally have all the equipment ready to send out yeasts from my yeast library to other (home)brewers. My yeast lab will mainly focus on different Brettanomyces strains, other souring bugs, blends and any kind of not commercially available yeast strains. To have a look at my preliminary yeast program please visit the respective site.
Please further notice that I don’t accept any orders. Currently, the strains are only available to collaborators for testing purposes. I plan to release strains on an irregular basis. All information concerning releases, costs, cell counts etc. are and/or will be available on the respective site.
Cheers, Sam
Beer Travel – Brussels Part 3
Eureka, and we already get to the last post about my latest trip to Brussels. If you haven’t read the previous two check out this page. Now, I only mentioned that the third post will be about a brewery resident in Brussels. And if you know me and Brussels well enough you already guessed the brewery. It is…”drum roll”… Cantillon. The pictures below are courtesy of my brother and most of the information are taken from the pamphlet you get for the brewery tour.
 Cantillon is one of the remaining traditional Lambic breweries in the world and is still a family business founded in 1900 by Paul Cantillon now within Brussels. They still use the old brewing equipment and brew an average of 1700 hectoliters (1450 barrels) of beer a year. In the 1960ies Jean-Pierre Van Roy carried on brewing and today Jean Van Roy is taking care of the legendary brewery. As we visited the brewery both Van Roy’s were present and Jean-Pierre Van Roy himself gave the brewery tour…
Cantillon is one of the remaining traditional Lambic breweries in the world and is still a family business founded in 1900 by Paul Cantillon now within Brussels. They still use the old brewing equipment and brew an average of 1700 hectoliters (1450 barrels) of beer a year. In the 1960ies Jean-Pierre Van Roy carried on brewing and today Jean Van Roy is taking care of the legendary brewery. As we visited the brewery both Van Roy’s were present and Jean-Pierre Van Roy himself gave the brewery tour…
The brewery is open for visits. You pay a small entry fee and get a nice 15 page pamphlet with some information about the brewing process and the products of Cantillon and two vouchers for a beer tasting after the tour. The brewery tour begins in the brewing area.
 For the mash they use 35% raw Belgian wheat and 65% malted barley. They mix the grains with water and hold the mash temperature for 2 hours and steadily rise the temperature from 42°C to 72°C. They then lauter with hot water and collect the wort in the hop boiler upstairs.
For the mash they use 35% raw Belgian wheat and 65% malted barley. They mix the grains with water and hold the mash temperature for 2 hours and steadily rise the temperature from 42°C to 72°C. They then lauter with hot water and collect the wort in the hop boiler upstairs. 
The hop boiler is made of copper and is heated by steam and has a propeller to mix the hops with the wort. They cook the wort for 3 to 4 hours with aged hops. During the boil they lose 2500 L due to evaporation from an original wort volume of 10000 L. After the primary fermentation the alcohol level is at approximately 5 ABV.  Next you climb up into the attic of the brewery. Up there they store their malt and wheat supplies and the hops. And there is the coolship were all the magic happens. After the boil the 7500 L of hot wort are pumped into the coolship to cool down. The large surface area helps to cool down the wort relatively fast. The cooling is done over night during the brewing season of October to April. During the cooling process a broad range of wild yeasts, bacteria etc. fall into the cooling wort. And these microorganisms lead to the spontaneous fermentation and in the end to the flavors in the finished product you either love or hate. It is mentioned in the pamphlet that over 100 different yeast strains, 27 acetic acid bacteria and 38 different lactic acid bacteria have been found in a single Lambic.
Next you climb up into the attic of the brewery. Up there they store their malt and wheat supplies and the hops. And there is the coolship were all the magic happens. After the boil the 7500 L of hot wort are pumped into the coolship to cool down. The large surface area helps to cool down the wort relatively fast. The cooling is done over night during the brewing season of October to April. During the cooling process a broad range of wild yeasts, bacteria etc. fall into the cooling wort. And these microorganisms lead to the spontaneous fermentation and in the end to the flavors in the finished product you either love or hate. It is mentioned in the pamphlet that over 100 different yeast strains, 27 acetic acid bacteria and 38 different lactic acid bacteria have been found in a single Lambic.
In the morning, after the wort cooled down to 18 to 20°C the wort is transferred into a stainless steel fermentation vat for the primary fermentation.  After the primary fermentation the beer gets transferred into oak or chestnut casks.
After the primary fermentation the beer gets transferred into oak or chestnut casks.

 Cantillon use casks which have been used by wine makers or Cognac producers. Once in the barrels the fermentation continues and a lot of foam and carbon dioxide escapes from the barrel as it can easily be seen in some of the pictures above. There was a lot of hissing noise in this room with the barrels above and a very nice smell in the air. The next pictures shows is more clearly…
Cantillon use casks which have been used by wine makers or Cognac producers. Once in the barrels the fermentation continues and a lot of foam and carbon dioxide escapes from the barrel as it can easily be seen in some of the pictures above. There was a lot of hissing noise in this room with the barrels above and a very nice smell in the air. The next pictures shows is more clearly…
 The vigorous fermentation slows down after some weeks and the barrels can be sealed. Now begins the maturing process which can take years.
The vigorous fermentation slows down after some weeks and the barrels can be sealed. Now begins the maturing process which can take years.
 Lambic is beer straight from the barrel and thus contains no carbonation at all. On the other hand, Gueuze is a blend of Lambics of different ages. Due to remaining sugars in the young Lambic (1-year-old) the fermentation in the Gueuze goes on and provides carbon dioxide to carbonate the beer. The 3-year-old Lambic provides the taste in the Gueuze. Yet another product of Cantillon are fruit beers. Lambics with fruits such as sour cherries, raspberries, grapes or apricots. The fruits (150 kg) are added to 500 L of 2-year old Lambic during the summer season (July to August). The fruits then stay in the Lambic for at least another three-month before the fruit beer is bottled. For bottling young Lambic is added again to supply the carbonation in the bottle. Lambics with sour cherries are called Kriek, with raspberries Framboise and the ones with grapes and apricots are somewhat specific for Cantillon. Vigneronne for the one with grapes and Fou’ Foune with apricots.
Lambic is beer straight from the barrel and thus contains no carbonation at all. On the other hand, Gueuze is a blend of Lambics of different ages. Due to remaining sugars in the young Lambic (1-year-old) the fermentation in the Gueuze goes on and provides carbon dioxide to carbonate the beer. The 3-year-old Lambic provides the taste in the Gueuze. Yet another product of Cantillon are fruit beers. Lambics with fruits such as sour cherries, raspberries, grapes or apricots. The fruits (150 kg) are added to 500 L of 2-year old Lambic during the summer season (July to August). The fruits then stay in the Lambic for at least another three-month before the fruit beer is bottled. For bottling young Lambic is added again to supply the carbonation in the bottle. Lambics with sour cherries are called Kriek, with raspberries Framboise and the ones with grapes and apricots are somewhat specific for Cantillon. Vigneronne for the one with grapes and Fou’ Foune with apricots.
Lambic is not made within some weeks like many top fermented beers. Patience is needed like stated by the sign below. Freely translated as “Time doesn’t respect those doing without it”
 And if you heard the story about the cobwebs in a Lambic brewery before, this is definitely true for Cantillon. There is this story that a lot of insects might harm the maturing beer and instead of using insecticides a lot of the brewers rely on spiders taking care of these insects. And thus Lambic brewers leave any cobwebs intact. However, have a closer look at the following barrel…
And if you heard the story about the cobwebs in a Lambic brewery before, this is definitely true for Cantillon. There is this story that a lot of insects might harm the maturing beer and instead of using insecticides a lot of the brewers rely on spiders taking care of these insects. And thus Lambic brewers leave any cobwebs intact. However, have a closer look at the following barrel…
 It might be hard to see but there are some sort of maggots around the plug enjoying the dripping Lambic… Enough of weird stuff.
It might be hard to see but there are some sort of maggots around the plug enjoying the dripping Lambic… Enough of weird stuff.
The tour ended in the barrel cleaning room and the bottling facility. Interestingly, Cantillon steam treats all their barrels prior to refilling. Thus killing any of the micro flora in the barrel formed during the last fermentation.
After the beer is bottled, the bottles stay another few months in the brewery which can easily be seen as you begin your brewery tour walking by huge bottle piles.  After the amazing tour and a small talk with Jean-Pierre Van Roy it was time for a tasting.
After the amazing tour and a small talk with Jean-Pierre Van Roy it was time for a tasting.  – Lambic straight from a barrel: Very pale color, not a lot of funky or sour aroma in the nose. On the palate some minor Brettanomyces notes but not (very?) sour. I encountered this before as I tasted a Lambic brewed by Girardin and was surprised that the Lambic after spending a year in the barrel is not sour.
– Lambic straight from a barrel: Very pale color, not a lot of funky or sour aroma in the nose. On the palate some minor Brettanomyces notes but not (very?) sour. I encountered this before as I tasted a Lambic brewed by Girardin and was surprised that the Lambic after spending a year in the barrel is not sour.
– Gueuze: Blend of Lambics. Pours with a pale color as well (picture above, glass on the very left side). In the nose the beautiful character one expects from a Gueuze. Lactic and acetic sourness, lots of barnyard, earthiness and some leathery smell as well. On the palate a decent sourness combined again with a funky earthiness. What a treat!
– Kriek: Lambic with sour cherries. Pours with a red color (glass on the right side in picture above). Some cherry character in the nose with the additional sour lactic vinegar notes. On the palate a lovely cherry bouquet complemented with a decent amount of sourness. Not sweet as modern Krieks are.
– Iris: Brewed with pale malt and spontaneously fermented and dry hopped with Saaz hops. Pours with a darker yellow color (glass in the middle in the picture above). I never had this beer before and was quite amazed how awesome the Saaz hops work with a sour beer like this. It gives the beer a very nice spiciness. The Saaz hops are definitely detectable on the palate as well. Fantastic!
Luckily for me I already had a lot of the Cantillon beers before visiting the brewery. The only ones still missing are the Vigneronne (the Lambic brewed with grapes), the Saint-Lamvinus (Lambic brewed with Merlot grapes), the Fou’ Foune (Lambic brewed with apricots) and the Faro (Lambic blended with caramel and candy sugar). Unfortunately, none of the beers above were available at the brewery shop expect from one, the Fou’ Foune. So I went home with a 0.75 L bottle of Fou’ Foune and wait for an opportunity to open the bottle… Luckily the bottle survived the flight back home.
I know I am very lucky to get Cantillon beers in the first place and to have the opportunity to pay a visit to the brewery. Anyway, as I walked through the storage space of the brewery I saw three euro-pallets like the one below:
 Maybe there is your next Cantillon bottle in there somewhere? I hope so.
Maybe there is your next Cantillon bottle in there somewhere? I hope so.
I am very happy about the visit and will definitely go there for another visit in the future. The beers they brew are just amazing, although some of the visitors had a funny face expression as they lowered their noses in the glasses for the very first time. Cantillon is a definite must if you ever are in/around Brussels. Even if you are not into the whole Lambic/Gueuze stuff. It is an experience and who knows maybe some day you really like to drink a traditional Gueuze, Lambic or fruit Lambic as well. The only thing I would love to do is take some agar plates with me next time… Thanks for reading and stay tuned for further posts.